An Easy Way to Add NMS to YOLOv8 graph
A journey to seamlessly incorporate NMS into YOLOv8 graph, streamlining the inference process and simplifying your workflow.
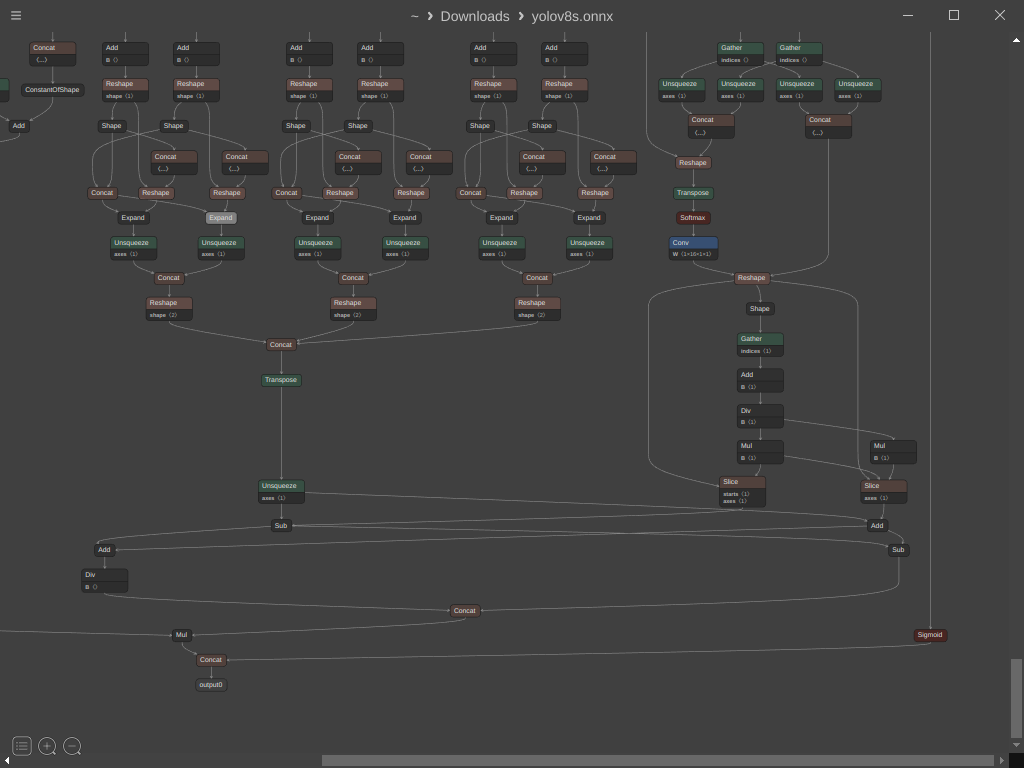
In the rapidly advancing realm of computer vision, object detection holds a crucial role in a wide array of applications, spanning from autonomous vehicles and surveillance systems to the intricate field of medical imaging. Amidst the plethora of object detection frameworks available, YOLO (You Only Look Once) has garnered immense popularity, celebrated for its real-time capabilities and precision. YOLOv8, the latest evolution in the YOLO lineage, introduces substantial improvements in object detection performance. However, it presents a challenge: the absence of built-in Non-Maximum Suppression (NMS) within the graph necessitates manual integration as part of your post-processing code.
In this article, we embark on a journey to seamlessly incorporate NMS into the graph, streamlining the inference process and simplifying your workflow.
You can find the code on GitHub
First of all, we need to understand the essential operations that we are going to use from `onnx.helper`
make_node
Used to create a processing node that will be added later to the graph.
which takes as arguments
op_type (string) – The name of the operator to construct
inputs (list of string) – list of input names
outputs (list of string) – list of output names
name (string, default None) – optional unique identifier for NodeProto
doc_string (string, default None) – optional documentation string for NodeProto
domain (string, default None) – optional domain for NodeProto. If it’s None, we will just use default domain (which is empty)
**kwargs (dict) – the attributes of the node.
example
transpose_bboxes_node = onnx.helper.make_node("Transpose", inputs=["/model.22/Mul_2_output_0"], outputs=["bboxes"], perm=(0, 2, 1))
make_tensor
Used to create a tensor with specified arguments
which takes as arguments
example
score_threshold = onnx.helper.make_tensor("score_threshold", onnx.TensorProto.FLOAT, [], [0.25])
make_tensor_value_info
Used to create a placeholder for a tensor
which takes as arguments
name (string) – tensor name
elem_type (int) a value such as onnx.TensorProto.FLOAT
shape (List of string or integer) optional
doc_string (string) optional
shape_denotation (List of string) optional
example
sigmoid_node = onnx.helper.make_tensor_value_info("/model.22/Sigmoid_output_0", onnx.TensorProto.FLOAT, shape=["batch", 80, 8400])
Now that we've familiarized ourselves with the essential operations to be employed, let's begin by the imports part and then utilizing a function to read the YOLOv8 ONNX file.
import onnx
import torch
import onnxsim
import onnxruntime as ort
import numpy as np
from torch import nn
from onnx.tools import update_model_dims
from onnx.compose import merge_models
from onnx.version_converter import convert_versiondef load_model(path):
onnx_model = onnx.load_model(path)
print(f"{path} Loaded")
onnx.checker.check_model(onnx_model)
return onnx_model
onnx_model = load_model("best.onnx")
graph = onnx_model.graphNow, we are going to define 3 tensors for score threshold, IOU threshold and maximum number of bboxes per class
score_threshold = onnx.helper.make_tensor(
"score_threshold",
onnx.TensorProto.FLOAT,
[],
[0.25])
iou_threshold = onnx.helper.make_tensor(
"iou_threshold",
onnx.TensorProto.FLOAT,
[],
[0.45])
max_output_boxes_per_class = onnx.helper.make_tensor(
"max_output_boxes_per_class",
onnx.TensorProto.INT64,
[],
[300])
graph.initializer.append(score_threshold)
graph.initializer.append(iou_threshold)
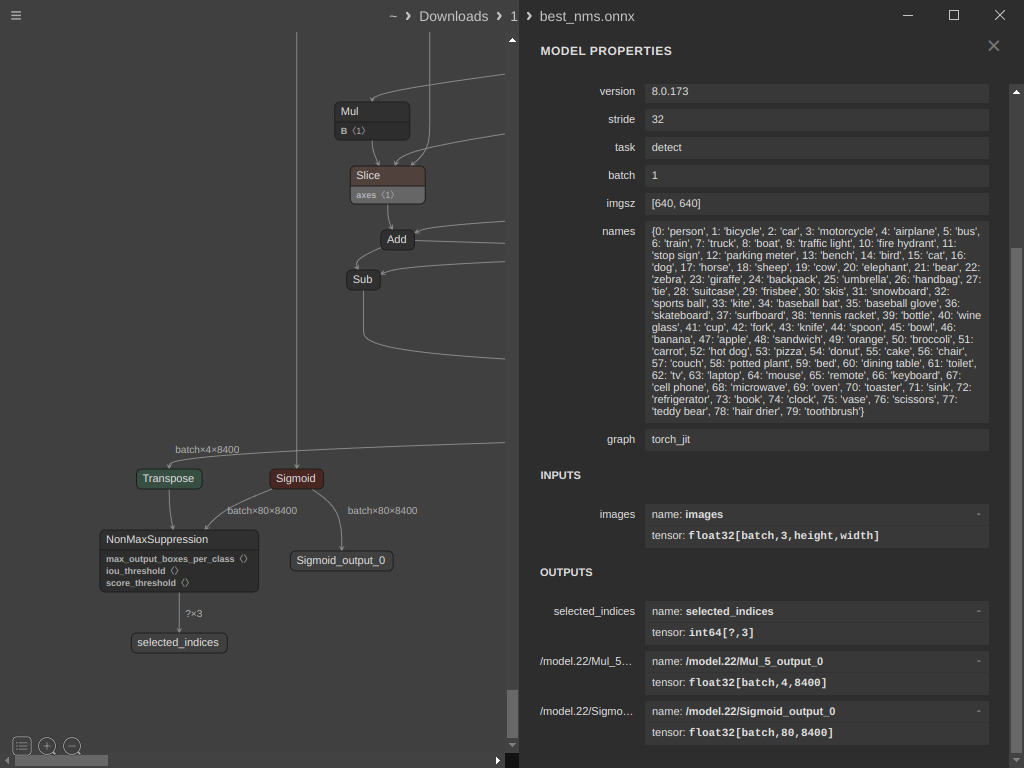
graph.initializer.append(max_output_boxes_per_class)After we initialized the previous tensors we need to get the names of the scores and boxes nodes from the graph, you can do this by opening the ONNX model using Netron, scroll down then select Mul node, Sigmoid node and Concat node
After selecting the Sigmoid node you can see the output name at the bottom right of the window which is “/model.22/Sigmoid_output_0“
Now lets define three variables that will hold the names of the nodes to make it easy to use later
Mul = "/model.22/Mul_5_output_0"
Sig = "/model.22/Sigmoid_output_0"
Con = "/model.22/Concat_25"Now lets create a node to transpose the boxes before passing it to the NMS and add it to the graph
transpose_bboxes_node = onnx.helper.make_node(
"Transpose",
inputs=[Mul],
outputs=["bboxes"],
perm=(0, 2, 1))
graph.node.append(transpose_bboxes_node)Now lets define the NMS node that will take the pre-defined tensors and the bboxes and sigmoid nodes.
Note that we need to set center_point_box=1 as we are dealing with PyTorch model that outputs [x_center, y_center, W, H] while the NMS node expect [x_min, y_min, x_max, y_max]
inputs = ['bboxes', Sig, 'max_output_boxes_per_class', 'iou_threshold', 'score_threshold']
outputs = ["selected_indices"]
nms_node = onnx.helper.make_node('NonMaxSuppression',
inputs,
outputs,
center_point_box=1)
graph.node.append(nms_node)Now let’s create a node for the selected indices which will hold [batch_index, class_index, box_index]
output_value_info = onnx.helper.make_tensor_value_info(
"selected_indices",
onnx.TensorProto.INT64,
shape=[None, 3])
graph.output.append(output_value_info)Next, we should remove the unused Concat node and the original output node from the model. Instead, we'll replace them with the Mul and Sigmoid nodes to create a model that provides the original boxes, confidence scores, and selected indices as output.
nc = 80 # Number of classes
last_concat_node = [
node for node in onnx_model.graph.node if node.name == Con
][0]
onnx_model.graph.node.remove(last_concat_node)
output0 = [
o for o in onnx_model.graph.output if o.name == "output0"
][0]
onnx_model.graph.output.remove(output0)
mul_node = onnx.helper.make_tensor_value_info(
Mul,
onnx.TensorProto.FLOAT,
shape=["batch", 4, 8400])
sig_node = onnx.helper.make_tensor_value_info(
Sig,
onnx.TensorProto.FLOAT,
shape=["batch", nc, 8400])
graph.output.append(mul_node)
graph.output.append(sig_node)Now Let’s save the model and check our progress
onnx.checker.check_model(onnx_model)
onnx.save(onnx_model, "best_nms.onnx")Our model now produces three values: selected indices, bounding box coordinates, and class information. However, we still need to process 8400 different records in post-processing and select from them using the specified indices. Can we address this challenge within the graph itself?
Indeed, we can. Let's explore how.
Initially, we need to create a transformation module that takes the selected indices, bounding boxes, and class information as input. This module will return only the selected boxes along with their corresponding class and associated scores.
class Transform(nn.Module):
def forward(self, idxTensor, boxes, scores):
batches = idxTensor[:, 0]
bbox_result = self.gather(boxes, idxTensor)
score_intermediate_result = self.gather(
scores,
idxTensor).max(axis=-1)
score_result = score_intermediate_result.values
classes_result = score_intermediate_result.indices
concatenated = torch.concat([
bbox_result[0],
score_result.T,
classes_result.T], -1)
return concatenated, batches
def gather(self, target, idxTensor):
pick_indices = idxTensor[..., -1:].repeat(1, target.shape[1])
if len(pick_indices.shape) == 2:
pick_indices = pick_indices.unsqueeze(0)
return torch.gather(target.permute(0, 2, 1), 1, pick_indices)As you can see this module outputs two tensors
The selected boxes, score, class with shape [number of boxes, 6]
The index of the box in the batch with shape [number of boxes]
Now we need to export this module to ONNX
# Use the nms model to simulate the input of the module
session = ort.InferenceSession("best_nms.onnx")
outname = [i.name for i in session.get_outputs()]
inname = [i.name for i in session.get_inputs()]
image = np.random.rand(8, 3, 640, 640).astype(np.float32)
output = session.run(outname, {inname[0]: image})
torch.onnx.export(Transform(), (
torch.tensor(output[0], dtype=torch.int64),
torch.Tensor(output[1]),
torch.Tensor(output[2])
),
"./NMS_after.onnx",
input_names=outname,
output_names=["det_bboxes", "batches"],
dynamic_axes={
"det_bboxes": {0: "num_results"},
"batches": {0: "num_results"},
})
nms_postprocess_onnx_model = onnx.load_model("./NMS_after.onnx")
nms_postprocess_onnx_model_sim, check = onnxsim.simplify(nms_postprocess_onnx_model)
onnx.save(nms_postprocess_onnx_model_sim, "./NMS_after_sim.onnx")After saving the model we have now two onnx files and we need to combine them in one file
best_nms.onnx which is the model + NMS
NMS_after_sim.onnx which is the transformation module
input_dims = {
"images": ["batch", 3, 640, 640],
}
output_dims = {
"selected_indices": ["batch", 3],
Mul: ["batch", "boxes", "num_anchors"],
Sig: ["batch", "classes", "num_anchors"],
}
target_ir_version = 18
updated_onnx_model = update_model_dims.update_inputs_outputs_dims(
onnx_model,
input_dims,
output_dims)
core_model = convert_version(updated_onnx_model, target_ir_version)
onnx.checker.check_model(core_model)
core_model.ir_version = 8
post_process_model = convert_version(
nms_postprocess_onnx_model_sim,
target_ir_version)
onnx.checker.check_model(post_process_model)
post_process_model.ir_version = 8
combined_onnx_model = merge_models(core_model, post_process_model, io_map=[
(Mul, Mul),
(Sig, Sig),
('selected_indices', 'selected_indices')
])
onnx.save(combined_onnx_model, './final_model.onnx')Now let’s try our final YOLOv8 with NMS model
Note that you may need to add the following code block to your post-process function, where boxes is the first output of your onnx model and indx is the second one.
final_result = [[] for _ in range(max(indx) + 1)]
for value, batch in zip(boxes, indx):
final_result[batch].append(value)Reference: Stitching non max suppression (NMS) to YOLOv8n on exported ONNX model